Phi Le
- Location: Worcester, Massachusetts
Data Scientist
Biostatistician and Bioinformatician
About Me
My full name Lê Long Phi in Vietnamese. I am from Vietnam. My hobbies when I have free time are watching action movies and reading.
Education
-
PhD in Applied Mathematics (2016): University of Missouri - Columbia.
-
MS in Biostatistics and Data Science (2020): University of Mississippi Medical Center
Employment
-
Data Scientist (Biostatistician, Bioinformatician, Machine Learning, Deep Learning): University of California - San Francisco: 2022-
-
Biostatistician: University of Mississippi Medical Center: 2018-2022
-
Mathematician: Syracuse University: 2016-2018
Recent Projects
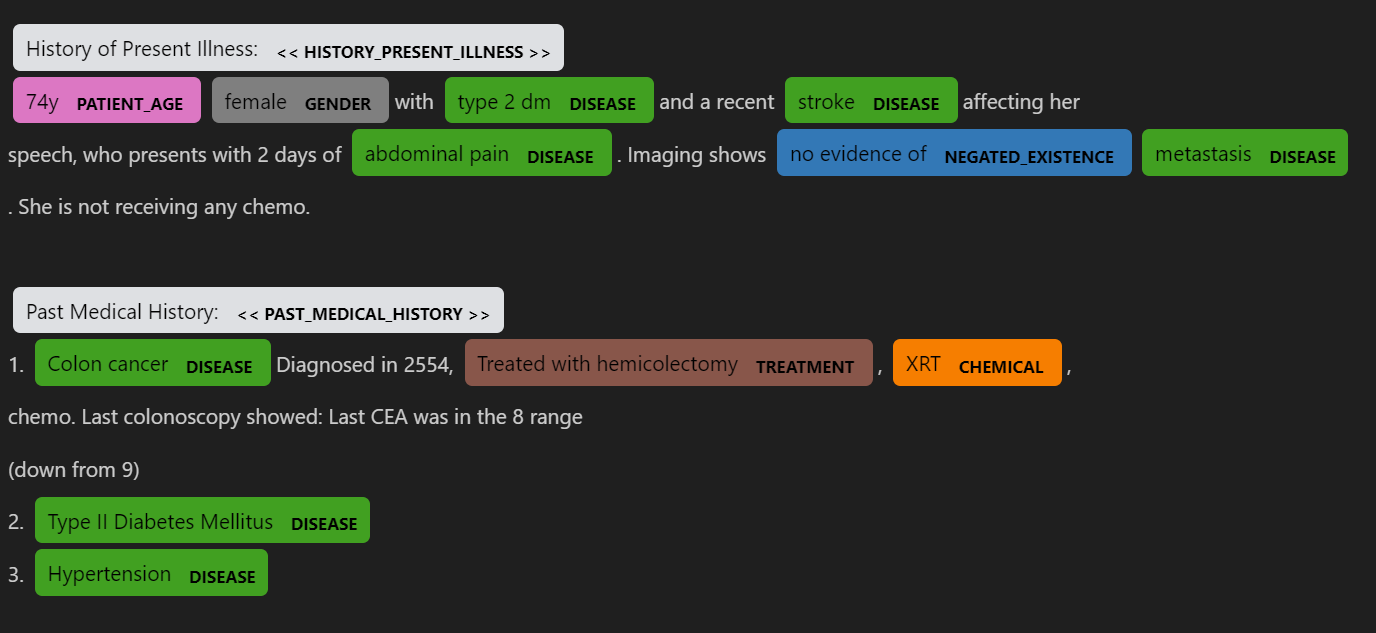
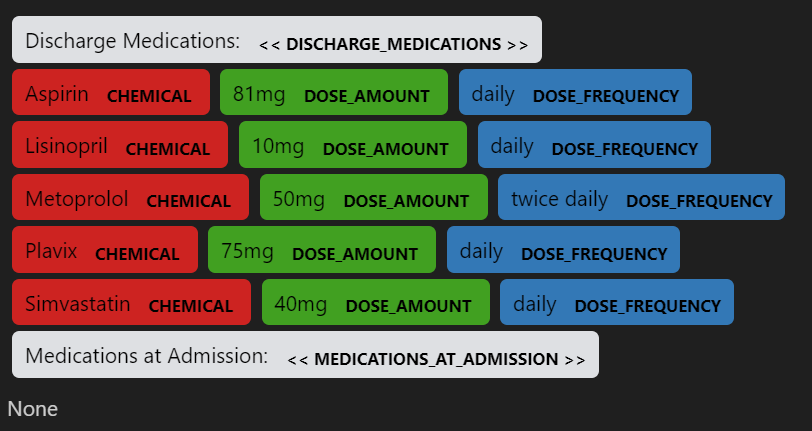
Using NLP to recognize medical information from physician's notes
I developed and modified Spacy, MedSpacy pipeline to do NLP analysis to extract needed information of patients
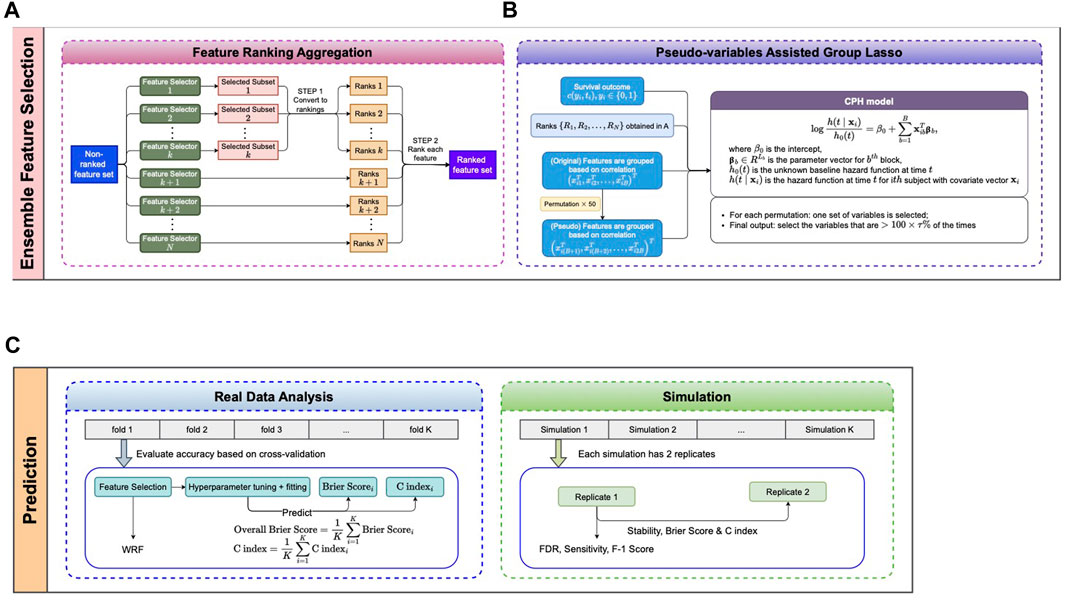
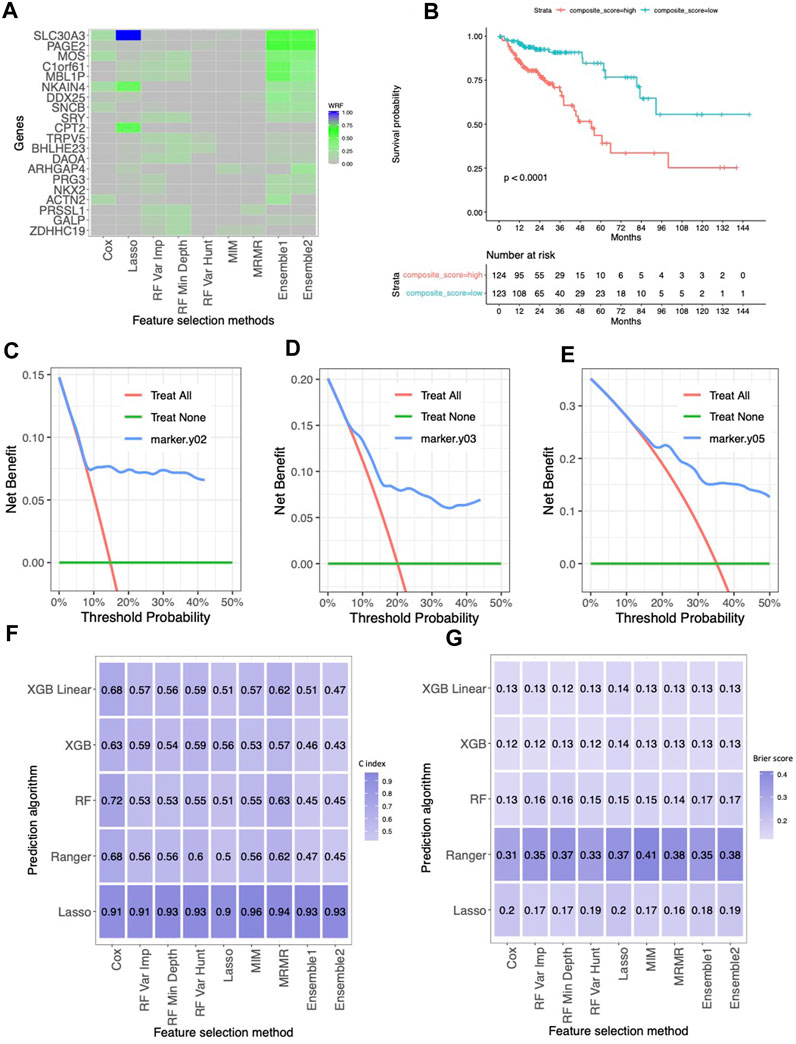
Feature selection model applied to gene expressions
We developed a novel ensemble method that invokes a Group Lasso Model with a permutation-assisted (PA) technique to find the feature associated with clinical outcomes of interest. Our model got high accuracy and less irrelevant selected features through various simulation scenarios due to the PA method. When using our selected features for the prediction model, our results showed better performance and robust predictions across different prediction models.
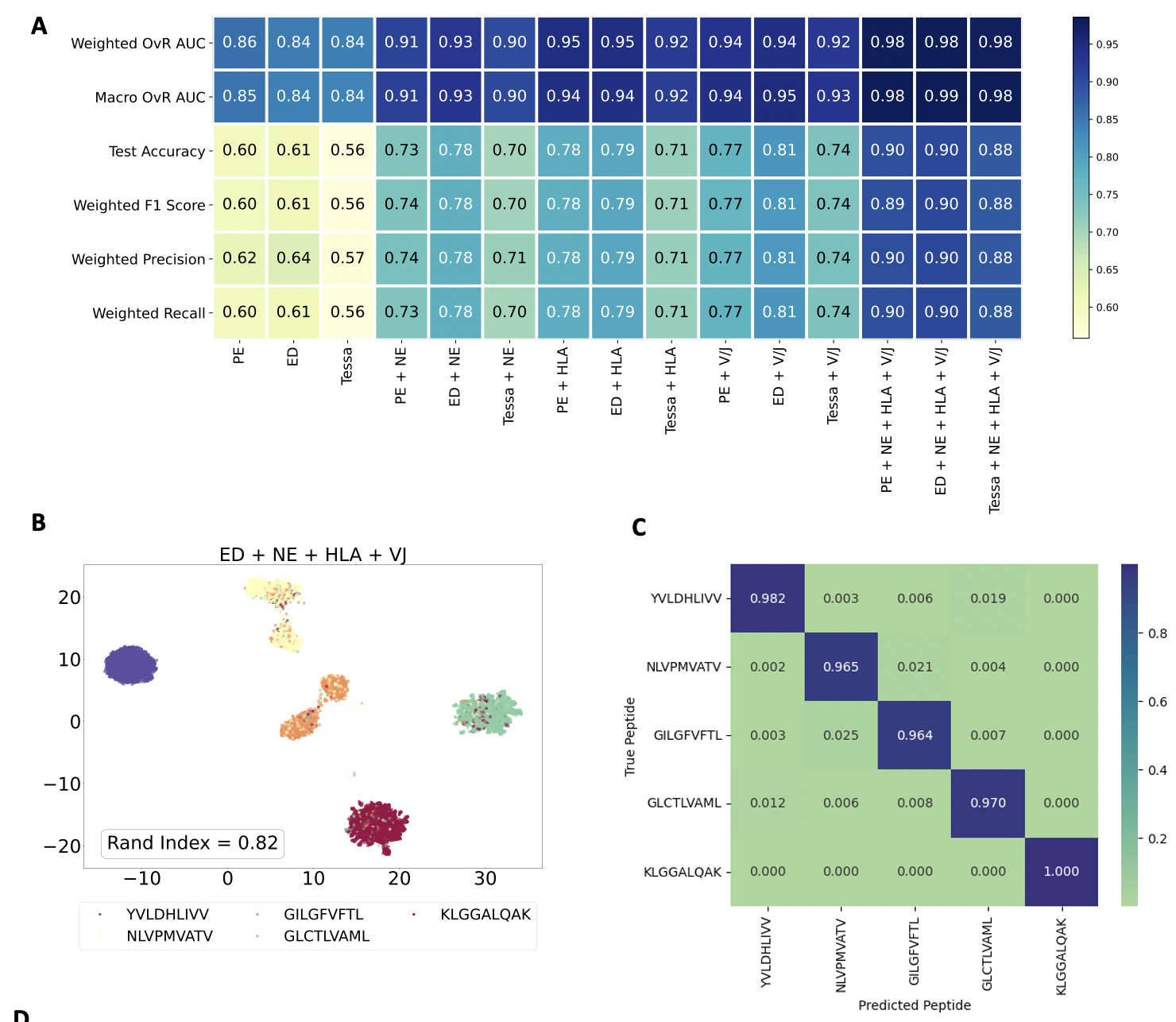
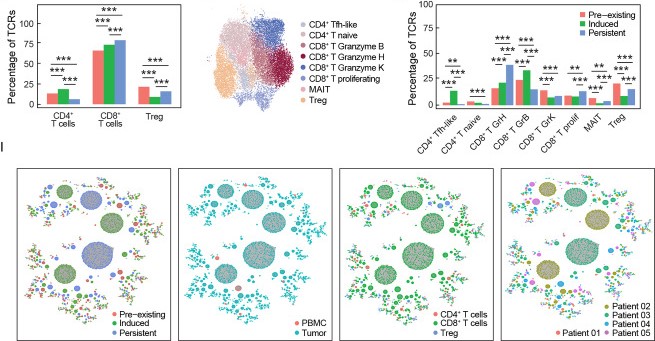
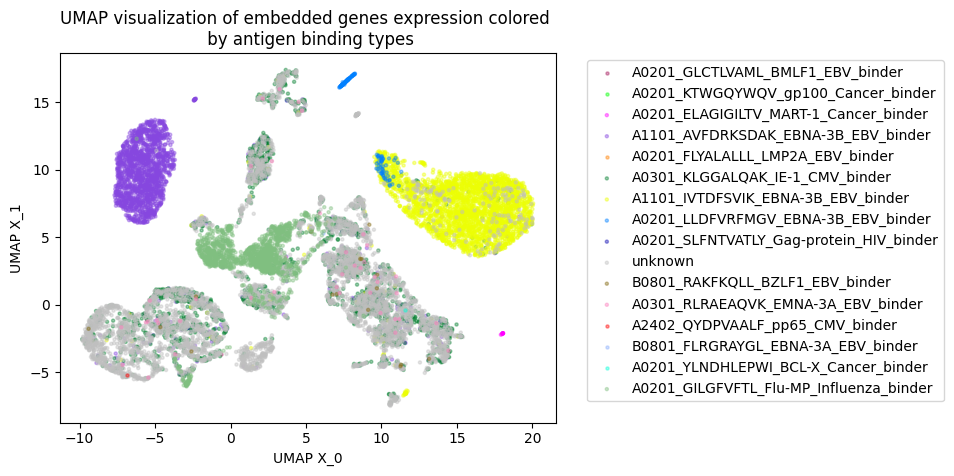
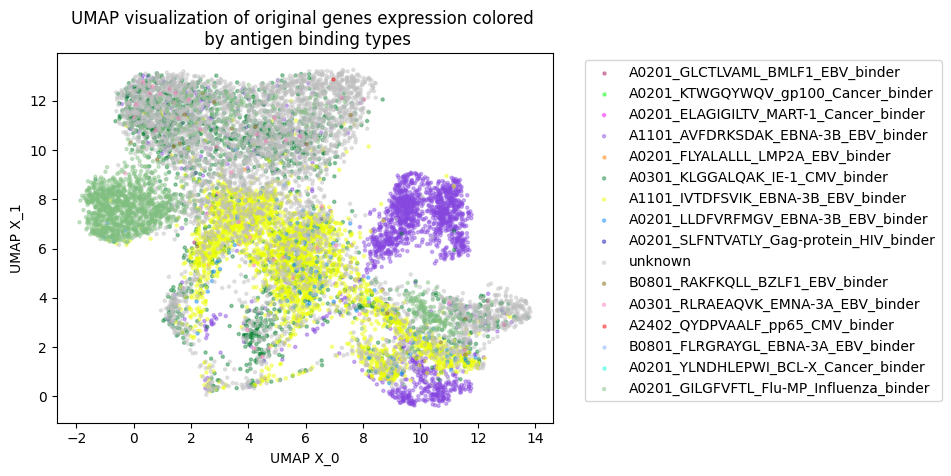
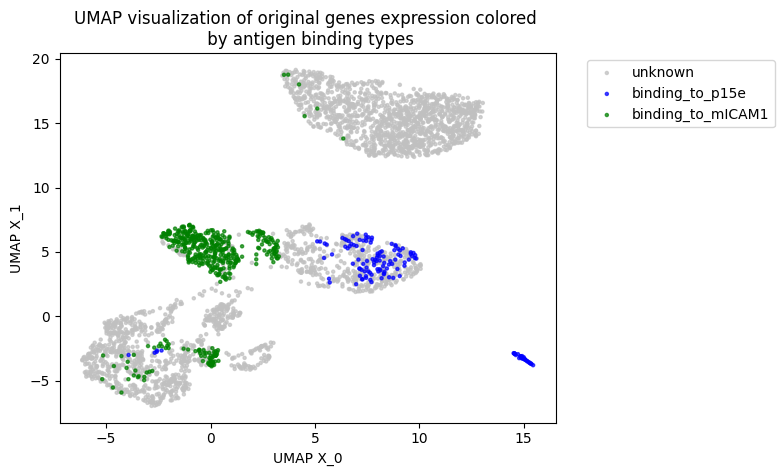
Tcell receptor - antigen peptide recognition prediction
Predicting the binding of T-cell receptors (TCRs) to peptides is essential for understanding the immune system and developing new immunotherapy treatments for diseases such as cancer. To achieve this, we developed various models using Deep Learning and Graph Neural Networks to encode letter-based amino acid sequences of TCRs into numerical values, which increases data variation. Additionally, we built a Bayesian classification model to obtain a high-performance model that can check the probability of binding between TCRs and a list of antigen peptides and provide uncertainty levels of predictions.
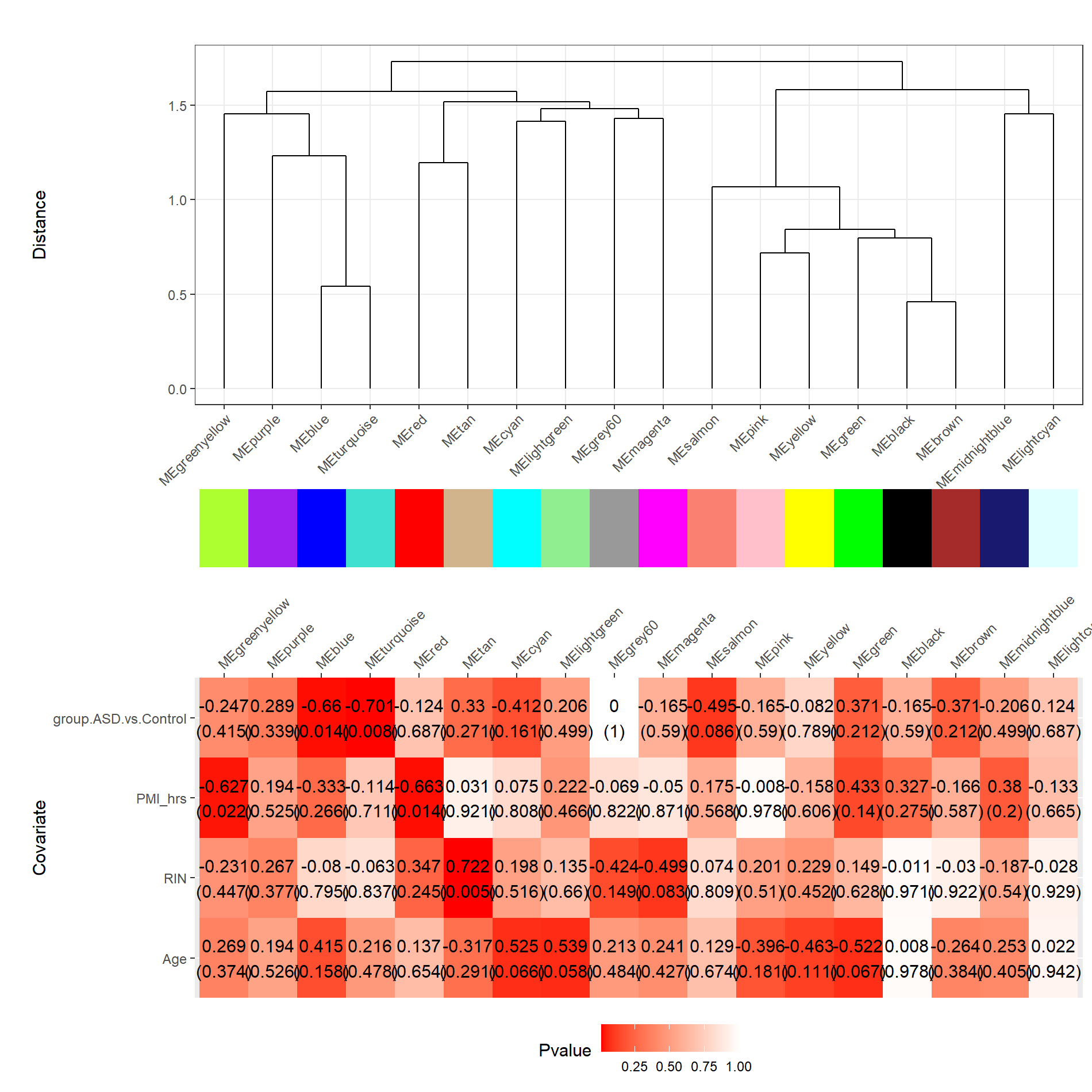
Gene clusters and their associations with ASD
I utilized WGCNA and DGE analyses to identify gene modules associated with Autism Spectrum Disorder (ASD) and discern significant gene expression differences compared to individuals without ASD.
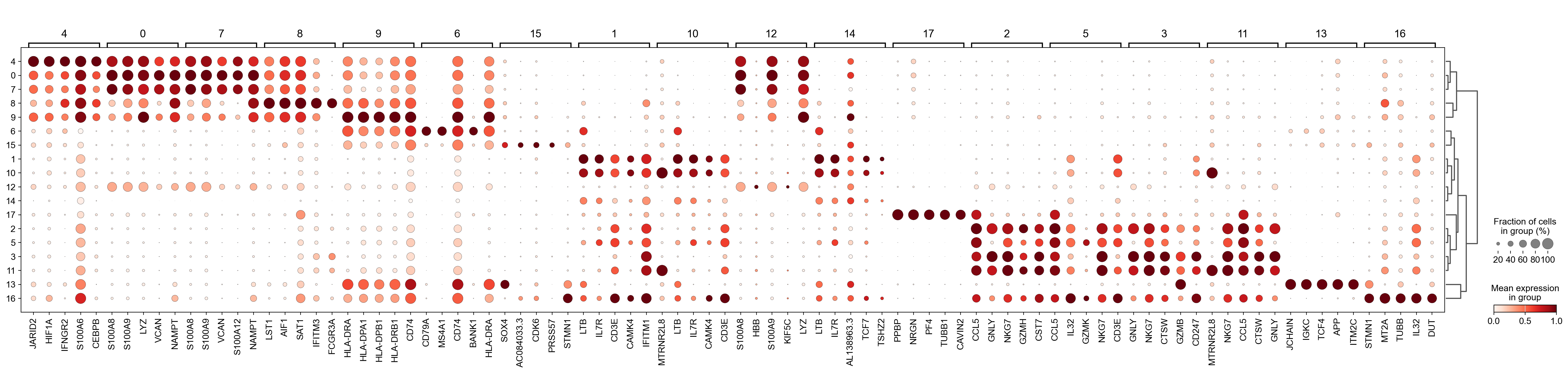
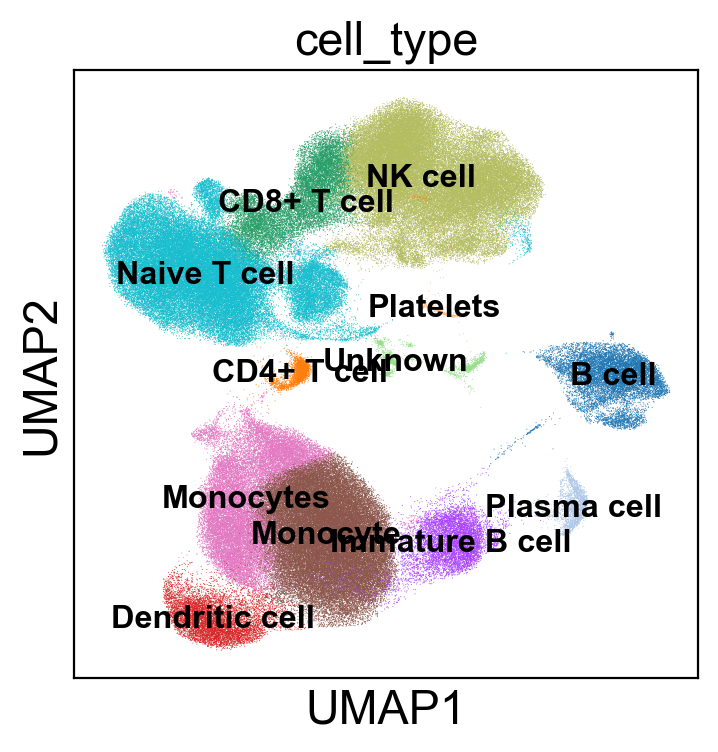
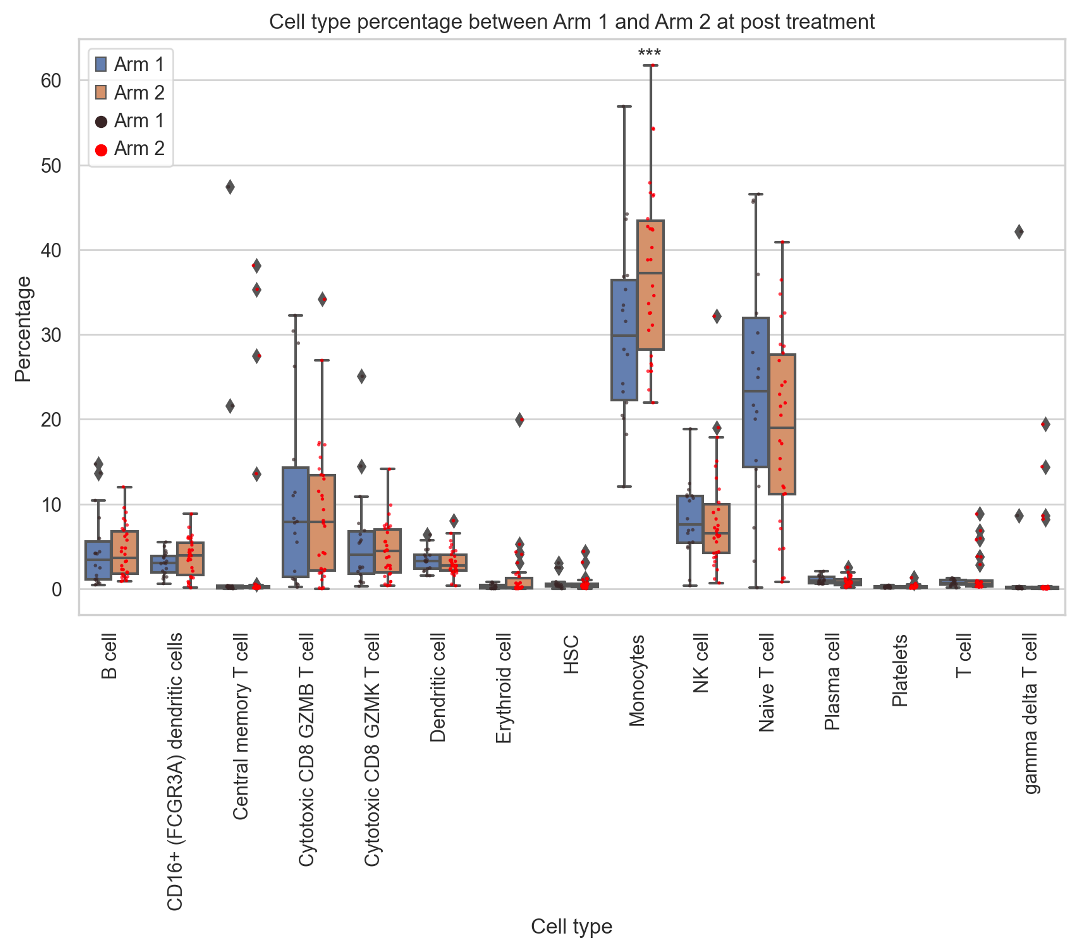
Single cell analysis pipeline
We developed a pipeline to conduct single-cell RNA-seq analysis from FASTQ files to annotate cells using marker genes. Additionally, I perform statistical analysis to examine changes in immune cells across different treatments and timepoints
Neoadjuvant CD40 Agonism Remodels the Tumor Immune Microenvironment in Locally Advanced Esophageal/Gastroesophageal Junction Cancer
In this project, I contributed to analyzing B-cell receptor similarities through network analysis and performed statistical analysis across different time points and features of B cells.
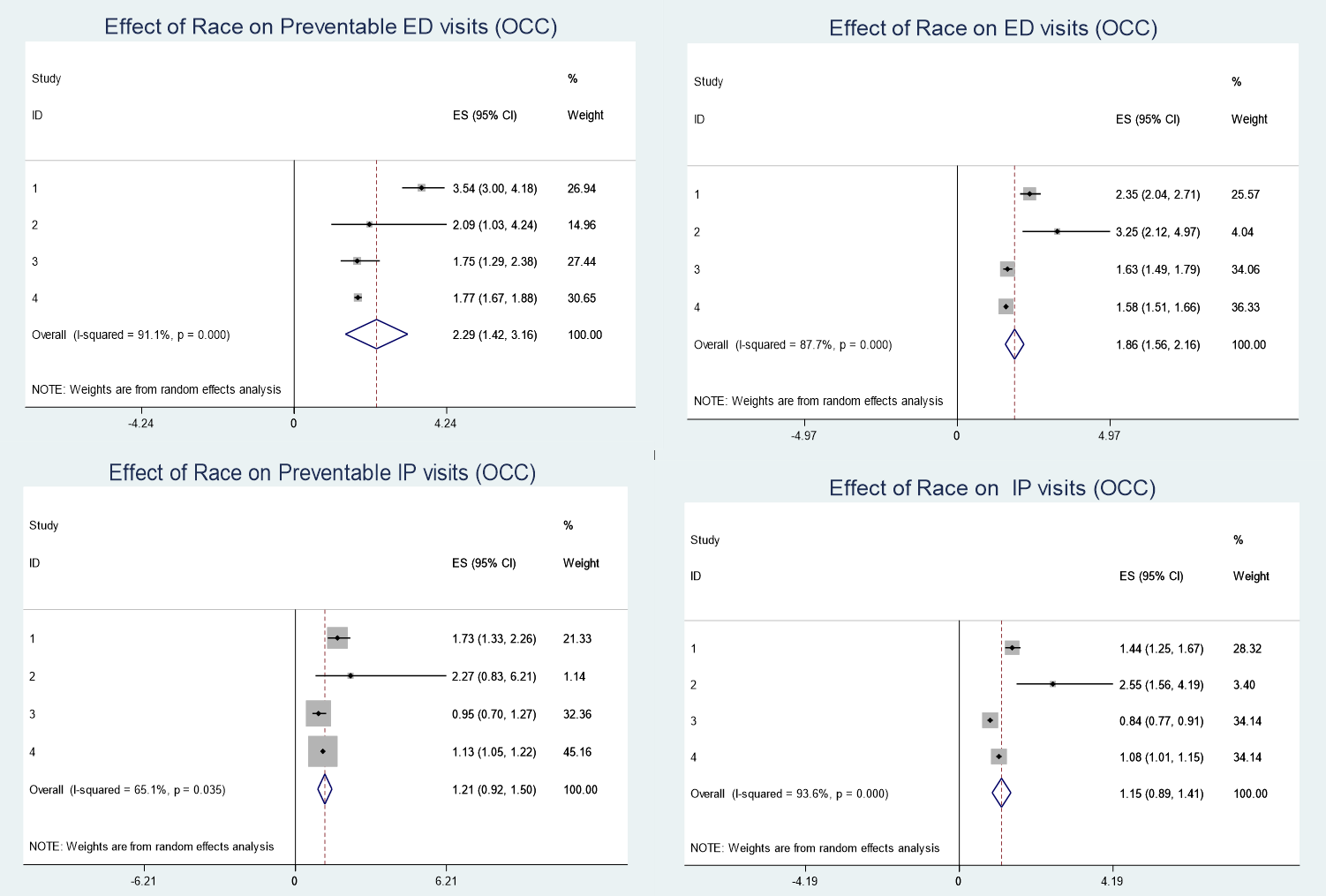
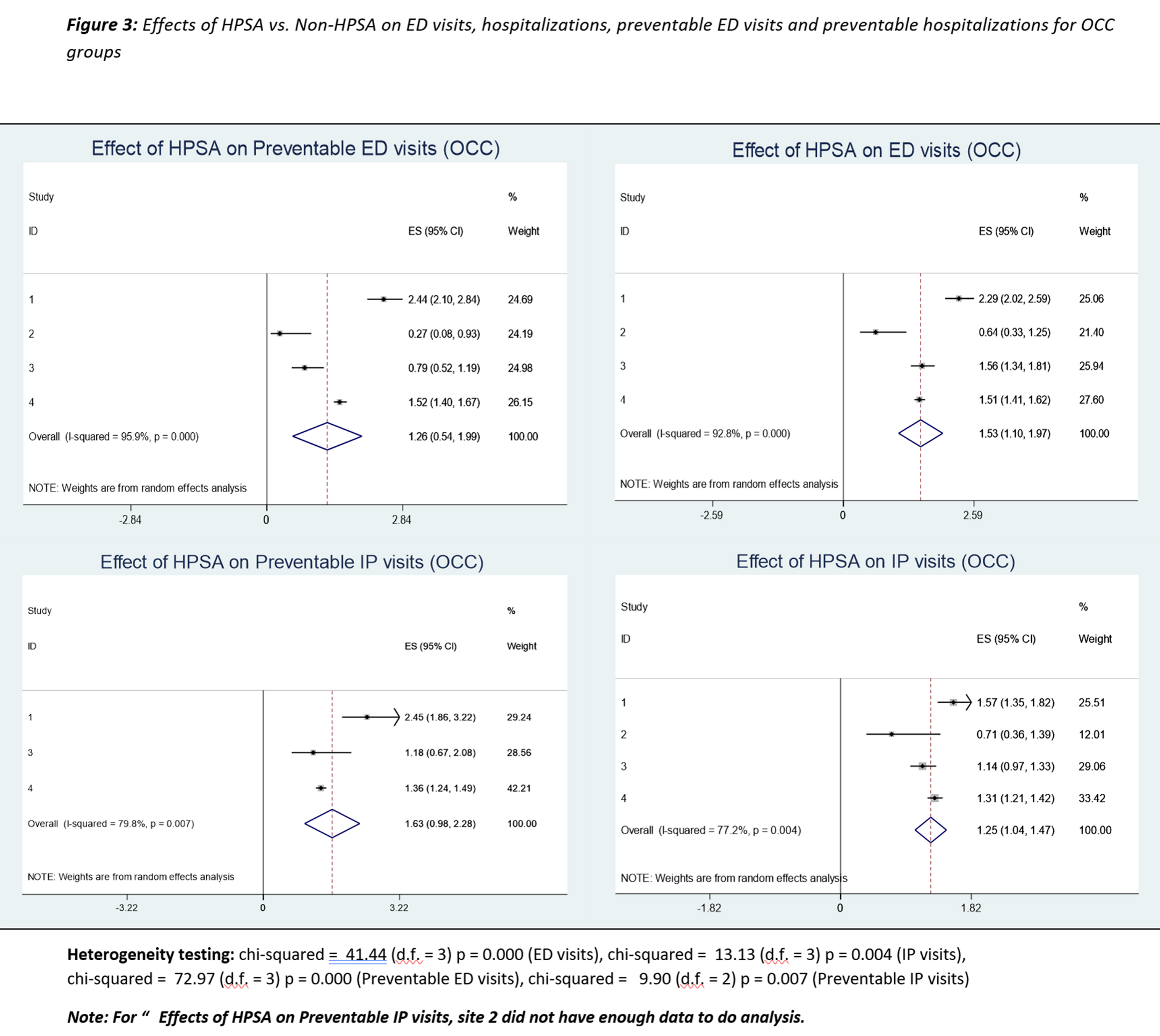
Effect of geographic and racial disparities on continuity of care and healthcare utilization among patients with obesity-associated chronic conditions
In this project, we apply Meta-analysis method involving practice-based research networks (PBRNs) for 3 large academic medical centers and one large integrated to study the Effect of geographic and racial disparities on continuity of care and healthcare utilization among patients with obesity-associated chronic conditions health care system in Tennessee, Mississippi, and Louisiana
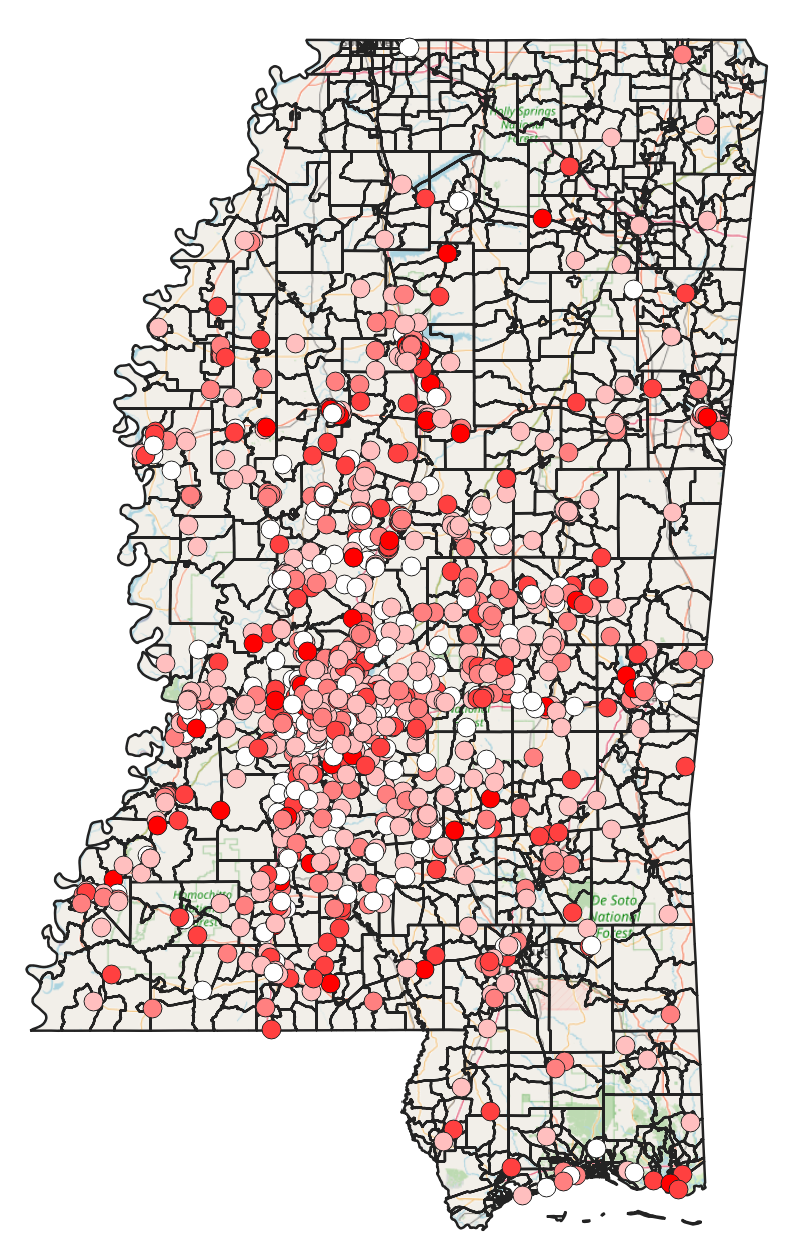
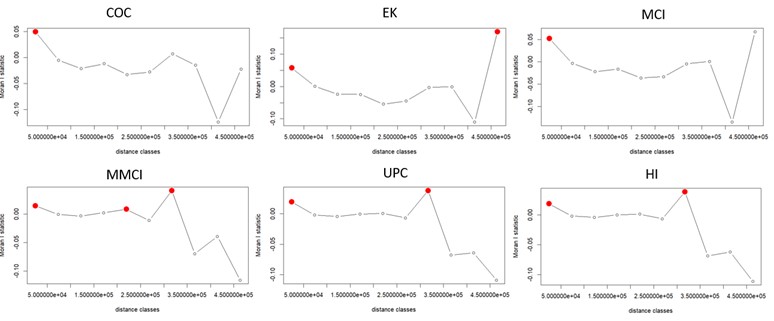
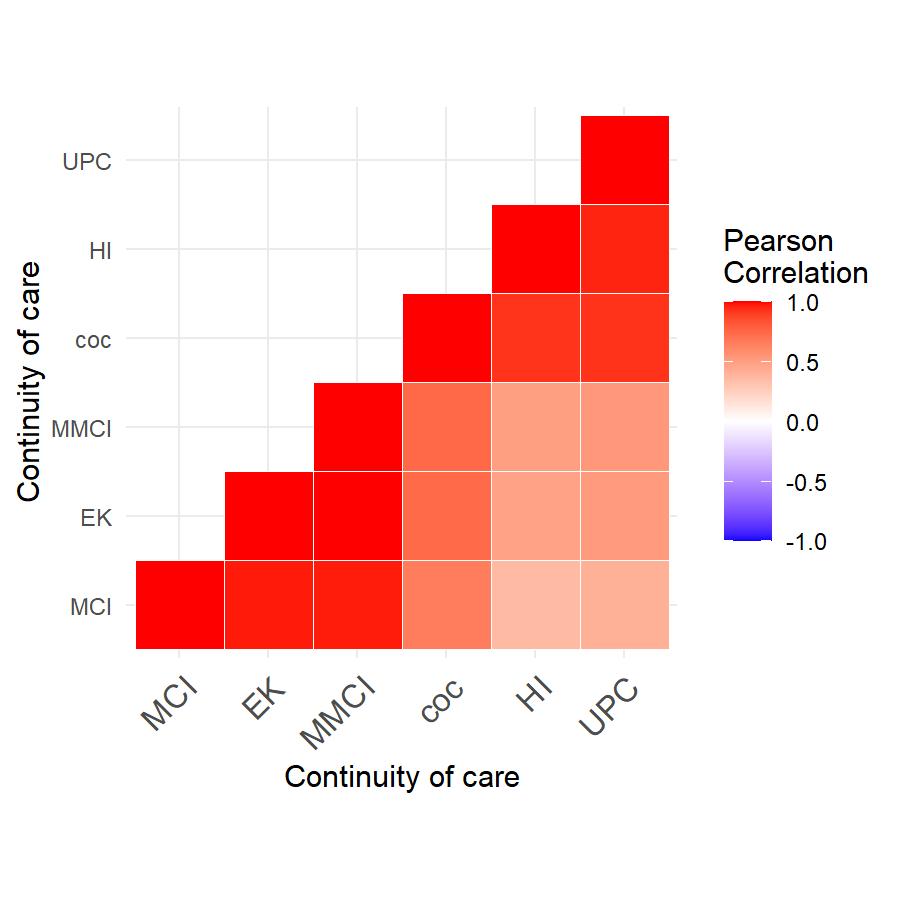
Spatial correlation and its effects on the continuity of care, length of stay at the hospital and readmission within 30 days
In our cohort study conducted in Mississippi, we examined whether increasing continuity of care provided protection to patients with chronic diseases against emergency visits and hospitalization readmissions. To investigate this, we employed spatial-temporal models that took into account the influence of geographic locations on the risk of experiencing emergency visits and hospitalizations. Notably, our approach diverged from classical linear regressions by incorporating the correlation effects of the neighborhood, which arise from Social Determinants of Health factors. This allowed for a more comprehensive analysis of the data and yielded insights into the potential benefits of improved continuity of care for patients with chronic diseases.
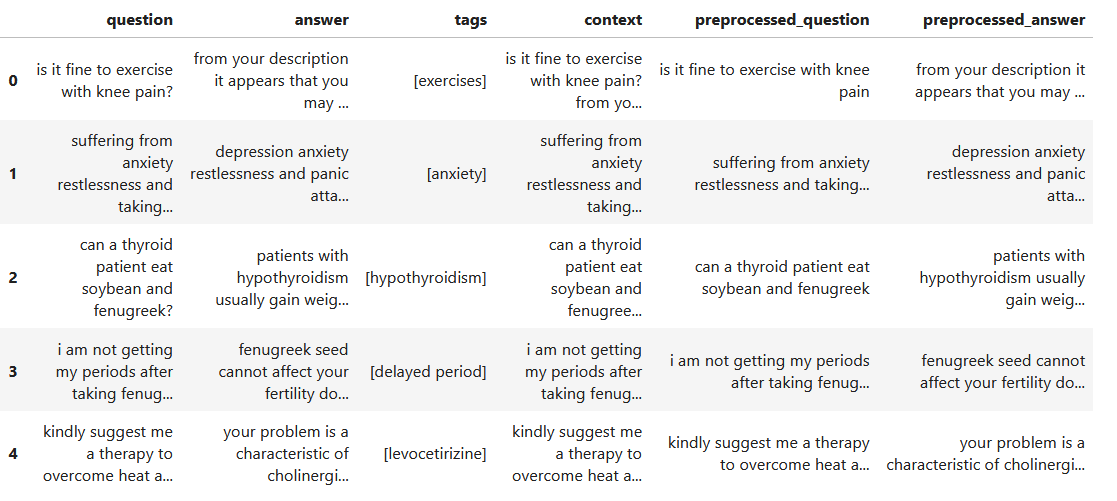
Medical tags from patients' descriptions of health questions
I utilize the BERT model to construct a natural language processing framework that automatically assigns tags to the specific health concerns inquired by users, based on their descriptions of medical issues.
Integration TCR and scRNA-seq sequences to clusters T-cells
By using GNN, we developed a novel model to integrate amino acid sequence based TCR and numerical value gene expression counts. Our results showed that we can be better cluster T-cells that can bind to the same antigen peptide from real data provided by 10X and mouse data
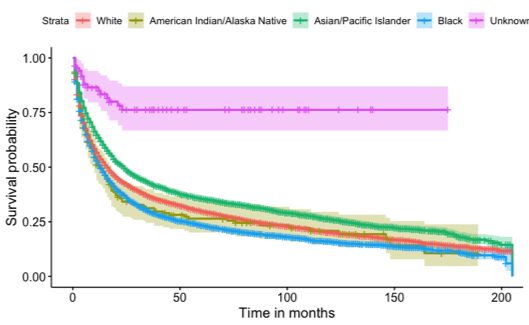
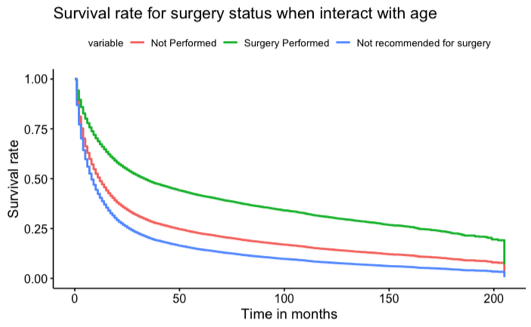
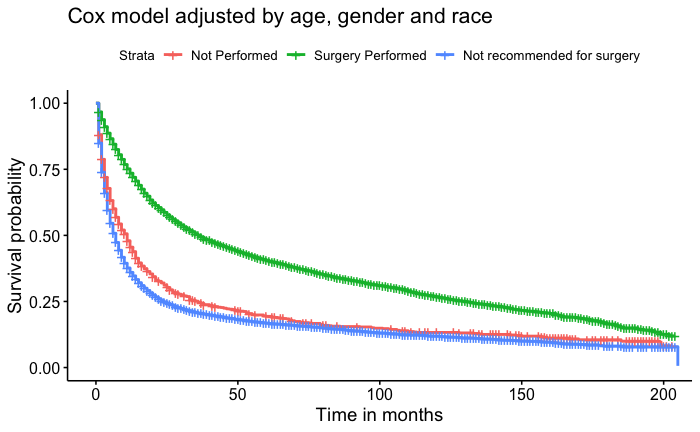
Some Observations of Gastric Cancer from SEER database
In this retrospective cohort survival study, we used the Cox proportional hazard model to investigate if undergoing surgery for non-related cancer issues would increase the lifespan of patients. In addition to our primary model, which was adjusted for age, race, and gender, we also conducted univariate models for age, gender, race, and surgery status, as well as a model for the interaction of surgery status and age. Our results showed that males had a 3% higher risk of death compared to females. Regarding race, the median survival times were 24 months for Asian/Pacific Islanders, 16 months for White individuals, 13 months for Black individuals, and the lowest at 12 months for American Indian/Native Alaskan individuals. We found conclusive evidence that surgery positively impacted patients' lifespans. Patients who underwent surgery had a median survival time of 36 months, while those who did not undergo surgery and were not recommended for surgery had median survival times of 11 months and 7 months, respectively.